The NovaSeq X Plus offers Illumina’s most advanced, powerful sequencing to date, surpassing the performance of the current workhorse, the NovasSeq 6000, while dramatically lowering costs. It features a 2.5-fold increase in output, lower cost per gigabase and per read, streamlined informatics, and sustainability advancement.
The new XLEAP-SBS chemistry delivers increased speed and greater fidelity. Ultrahigh-density patterned flow cells contain tens of billions of nanowells at fixed locations, a design that provides even spacing of sequencing clusters. Flow cells are named according to the billions (B) of clusters each are capable of producing, and the UMGC offers both the 10B and the 25B flow cells. Each flow cell contains 8 independent lanes, which can be used to load up to 8 separate library pools.
Highlighted applications
- Whole-genome or whole-transcriptome sequencing
- Whole-genome metagenomic sequencing
- Population-scale sequencing
Outsourcing
The UMGC has determined that for the time being, access to the NovaSeq X Plus will be both faster and less expensive when outsourced, and we have partnered with three academic cores to achieve this.
Output
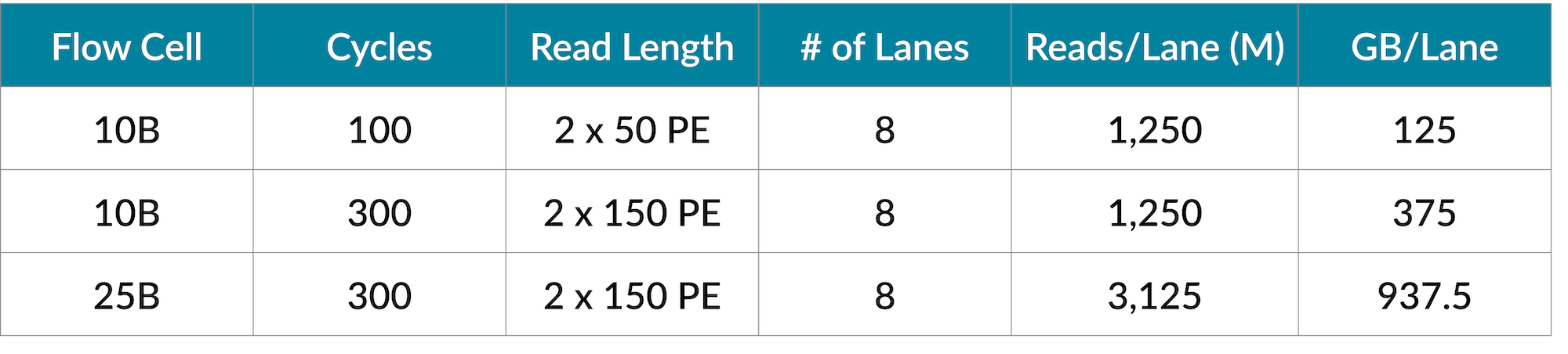
The 10B flow cell has options for either the 100-cycle (2 x 50 PE) or 300-cycle (2 x 150 PE) sequencing kits. The 25B flow cell is only available in a 300-cycle format (2 x 150 PE).
Benefits
- Affordably analyze whole genomes: Expand research with whole-genome sequencing
- Add statistical power: Manage bigger sample cohort sizes for population-scale studies.
- Sequence more deeply: Identify rare genetic events for cancer and genetic disease applications.
- Examine multiple dimensions: Combine data from genomes, transcriptomes, and epigenomes for a more vivid picture of biological systems.
- Transition to high-resolution approaches: Including single-cell sequencing and spatial analysis for insights into complex tissues.
Questions
Please contact Aaron Becker or Elyse Cooper at next-gen@umn.edu to receive a project proposal using the NovaSeq X Plus platform.
UMN Rates
High Output Sequencing Rates Summary
| 100-cycle (50 PE) | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| NovaSeq X+ 10B. 1,250 M reads/lane. | $1,355.97 | $2,000.34 | N/A |
| Turnaround time. | 10 to 15 days | 5 to 10 days | N/A |
| 300-cycle (150 PE) | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| NovaSeq X+ 10B. 1,250 M reads/lane. | $1,681.89 | $2,476.26 | N/A |
| Turnaround time. | 10 to 15 days | 5 to 10 days | N/A |
| NovaSeq X+ 25B. 3,250 M reads/lane. | $2,740.04 | $4,021.41 | N/A |
| Turnaround time. | 10 to 15 days | 5 to 10 days | N/A |
Notes
1. Library prep: Internal (UMN) users may either make their own libraries and submit them to the UMGC for QC and outsourced sequencing or have the UMGC generate libraries for them. External (non-UMN) users may only access outsourced sequencing using UMGC-created libraries.
2. Output: The above output/lane are specifications reported by Illumina. Actual output may vary by library type.
3. Tiers: Turnaround time (TAT) is in business days. TAT is for sequencing only and does not factor in the TAT for library prep or other companion services. More information about Pricing Tiers.
External Rates
High Output Sequencing Rates Summary
| 100-cycle (50 PE) | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| NovaSeq X+ 10B. 1,250 M reads/lane. | $1,947.17 | $2,752.03 | N/A |
| Turnaround time. | 10 to 15 days | 5 to 10 days | N/A |
| 300-cycle (150 PE) | Savings Tier | Speed Tier | Priority Tier |
|---|---|---|---|
| NovaSeq X+ 10B. 1,250 M reads/lane. | $2,415.19 | $3,408.63 | N/A |
| Turnaround time. | 10 to 15 days | 5 to 10 days | N/A |
| NovaSeq X+ 25B. 3,250 M reads/lane. | $3,934.71 | $5,535.16 | N/A |
| Turnaround time. | 10 to 15 days | 5 to 10 days | N/A |
Notes
1. Library prep: Internal (UMN) users may either make their own libraries and submit them to the UMGC for QC and outsourced sequencing or have the UMGC generate libraries for them. External (non-UMN) users may only access outsourced sequencing using UMGC-created libraries.
2. Output: The above output/lane are specifications reported by Illumina. Actual output may vary by library type.
3. Tiers: Turnaround time (TAT) is in business days. TAT is for sequencing only and does not factor in the TAT for library prep or other companion services. More information about Pricing Tiers.
Submission
How to order
Contact next-gen@umn.edu for a project proposal using the NovaSeq X Plus.
Samples can be dropped off at one of our campus locations
1-210 Cancer & Cardiovascular Research Building (Minneapolis campus)
20 Snyder Hall (St. Paul campus)
Samples can be shipped to the following address:
Please send the tracking information to next-gen@umn.edu.
UMN Genomics Center
ATTN: Nasia Mead
3510 Hopkins Place N.
Building 4 Suite W402
Oakdale, MN 55128
612-625-7736
Deliverables
Data Release
There are four options for transferring data from the UMGC to clients: 1) delivery to the Minnesota Supercomputing Institute’s (MSI) high-performance file system, 2) download from a secure website, 3) download with Globus, or 4) shipment on an external hard drive. Please indicate your data delivery preference when placing an order for sequencing.
1. MSI storage
Internal clients have their data released to MSI's Shared User Resource Facility Storage (SURFS). Delivered data will be located in the "data_delivery" folder in your group's folder on MSI's primary filesystem (home/GROUP/data_delivery/umgc). MSI does not charge for SURFS storage costs, but files expire and are removed one year after they've been delivered. Files should be copied to other MSI storage locations such as Tier2, Tier3, or your group's "shared" folder before they expire.
2. Web download
Internal clients that opt-out of MSI storage and external clients can download their data from a secure website using either a web browser or a command-line download tool, complete instructions are provided in an email from the UMGC. The client’s data is available for download for 30 days, after which the data will be removed from the data download website and the client takes responsibility for storing the data.
3. Globus
Internal and External clients can use Globus to download their data. This is the recommended method for external clients to download large datasets.
4. Hard drive
External clients may have data shipped on a hard drive purchased by the UMGC and invoiced to the client at a cost of $250 per hard drive.
Data Recovery
The UMGC archives most customer data for a year and some datasets are retained for 5 years or more. If you need a dataset re-delivered email a request to next-gen@umn.edu to initiate data recovery. The UMGC does not provide any guarantee that data can be successfully recovered from the archive.